- Get link
- X
- Other Apps
My thesis project is a computer program designed to predict the three-dimensional structure of proteins given only their amino acid sequence. This system is also the first of a family of computer.
Protein Structure Prediction From Amino Acid Chain Folding
46 Zeilen Prediction of Protein Folding Rates from Geometric Contact and Amino Acid Sequences.

Predict protein folding. 15 Zeilen trRosetta is an algorithm for fast and accurate de novo protein structure prediction. An arcane form of molecular origami its importance is hard to overstate. Predicting the structure is not as easy as determining the minimum energy conformation because that brings about problems on its own.
A protein structure prediction method must explore the space of possible protein structures which is astronomically large. Here we propose a distance-guided protein folding algorithm based on generalized descent direction named GDDfold which achieves effective structural perturbation and potential minimization in two stages. Predicting how these chains will fold into the intricate 3D structure of a protein is whats known as the protein folding.
The way a protein folds is dependant on the amino acids its made of and the environment that its in. But these steps cant predict a. Protein folding has been a grand challenge in biology for 50 years.
PROTEUS2- is a web server designed to support comprehensive protein structure prediction and structure-based annotation. ORION - is a web server for protein fold recognition and structure prediction using evolutionary hybrid profiles. Other projects such as Googles AlphaFold have generated tremendous recent excitement by using advances in artificial intelligence to predict a proteins structure.
This is an important topic in the field of proteomics and one of the grand challenges of molecular biology. These problems can be partially bypassed in comparative or homology modeling and fold recognition methods in which the search space is pruned by the assumption that the protein in question adopts a structure that is close to the experimentally determined structure of. As demonstrated by Levinthals paradox it would take longer than the age of the known universe to randomly enumerate all possible configurations of a typical protein before reaching the true 3D structure - yet proteins themselves fold spontaneously within milliseconds.
Advances in the prediction of the inter-residue distance for a protein sequence have increased the accuracy to predict the correct folds of proteins with distance information. If you are unfamiliar with the protein. For the modeling step a protein 3D structure can be directly obtained from the selected template by MODELLER and displayed with global and local quality model estimation.
Phyre Standard Mode Login for job manager batch processing Phyre alarm and other advanced options. The importance of protein folding prediction. PROTEUS2 accepts either single sequences for directed studies or multiple sequences for whole proteome annotation and predicts the secondary and if possible tertiary structure of the query proteins.
To do so these approaches parse enormous volumes of genomic data which contain the blueprint for protein sequences. The prediction server is available online at. Various databases such as PDB SCOP and HOMSTRAD can be mined to find an appropriate structural template.
Since a protein can shift between several intermediate. The PHYRE automatic fold recognition server for predicting the structure andor function of your protein sequence. A parallel neural network predicted the angles of the joints between consecutive amino acids in the folded protein chain.
This folding is a result of amino acids coming together through attraction across disparate. The correlation between folding rates and protein sizes is not as large however. The topic of my masters thesis project at George Mason University is the study of a genetic algorithm approach to predicting protein structure in abstract proteins folded in 3-dimensional lattices.
Our method showed an excellent correlation of 096 between predicted and experimental folding rates of proteins. PredictProtein - Protein Sequence Analysis Prediction of Structural and Functional Features. A web server has been set up to predict the protein folding rates which takes the amino acid sequence and structural class information as input and displays the folding rate of the protein along with amino acid composition in the output.
The reason there is yet to be a perfected solution to the protein folding problem is because of all the possible conformations the protein could form. Most biological processes revolve around proteins and. Proteins are both the engines and the building blocks of all living things thus an understanding of their structure and behavior is essential to understanding how living things operate.
Alphafold Using Ai For Scientific Discovery Deepmind
Alphafold A Solution To A 50 Year Old Grand Challenge In Biology Deepmind
 The Game Has Changed Ai Triumphs At Solving Protein Structures Science Aaas
The Game Has Changed Ai Triumphs At Solving Protein Structures Science Aaas
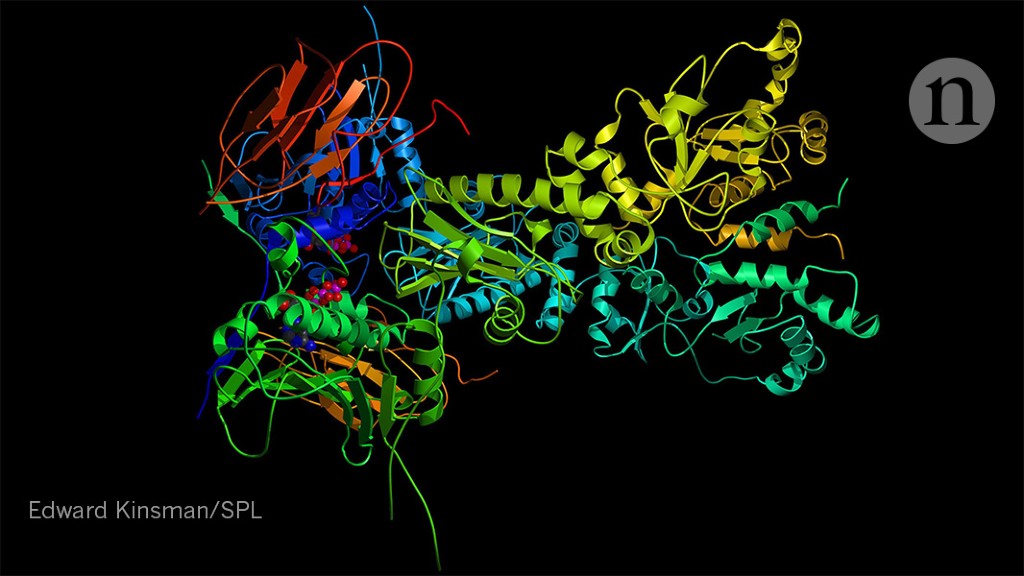 Ai Protein Folding Algorithms Solve Structures Faster Than Ever
Ai Protein Folding Algorithms Solve Structures Faster Than Ever
 Computational Pipeline For Protein Folding The Msa For The Protein Download Scientific Diagram
Computational Pipeline For Protein Folding The Msa For The Protein Download Scientific Diagram
/cdn.vox-cdn.com/uploads/chorus_image/image/68052989/global_distance_result.0.png) Go Watch This Video About An Ai System That Can Predict How Proteins Fold The Verge
Go Watch This Video About An Ai System That Can Predict How Proteins Fold The Verge
 Improved Protein Structure Prediction Using Predicted Interresidue Orientations Pnas
Improved Protein Structure Prediction Using Predicted Interresidue Orientations Pnas
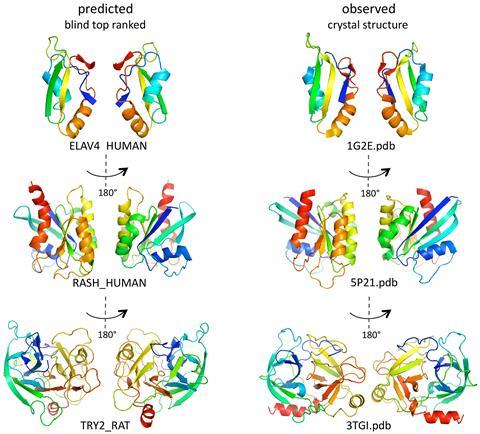 Go With The Fold Feature Chemistry World
Go With The Fold Feature Chemistry World
 A Guide For Protein Structure Prediction Methods And Software By Helene Perrin Medium
A Guide For Protein Structure Prediction Methods And Software By Helene Perrin Medium
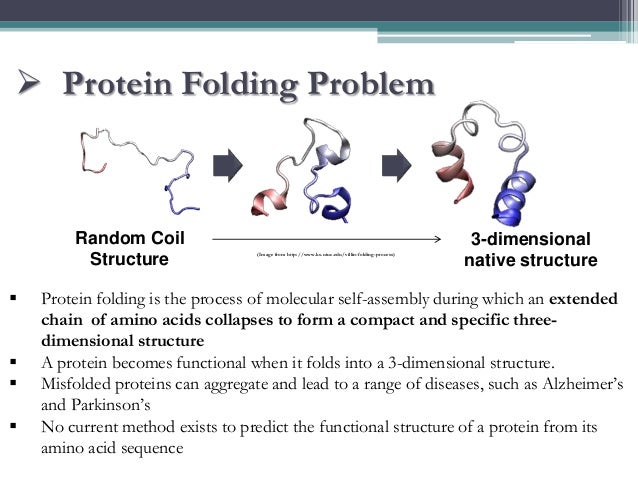 Protein Structure Prediction From Amino Acid Chain Folding
Protein Structure Prediction From Amino Acid Chain Folding
 Deepmind Ai Makes Breakthrough With Protein Folding Problem The New Stack
Deepmind Ai Makes Breakthrough With Protein Folding Problem The New Stack
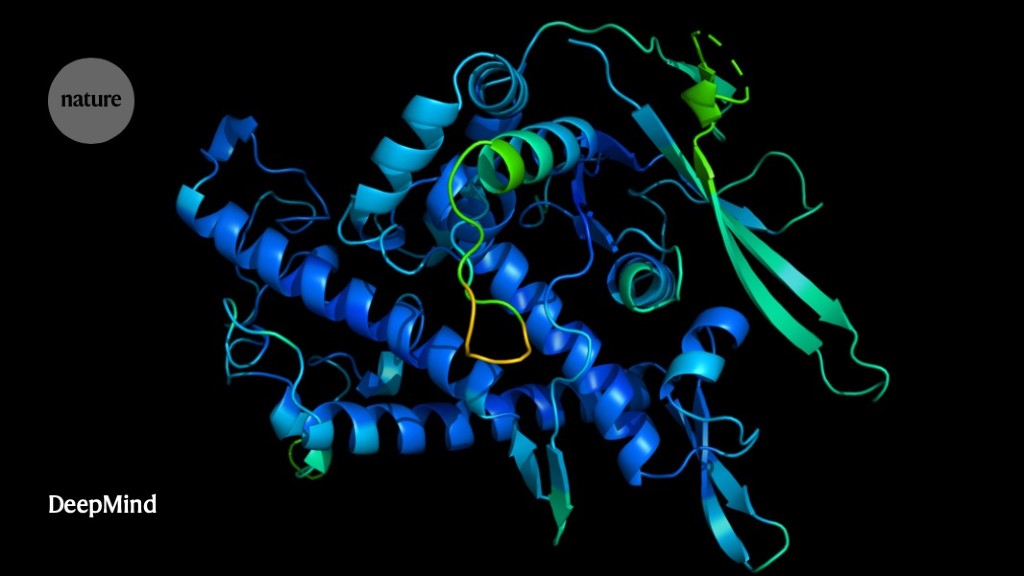 It Will Change Everything Deepmind S Ai Makes Gigantic Leap In Solving Protein Structures
It Will Change Everything Deepmind S Ai Makes Gigantic Leap In Solving Protein Structures


Comments
Post a Comment